

The Monte Carlo simulation was performed by ARCHER-MIC and GPU codes, which were benchmarked against a modified parallel DPM code. The phantom was generated from DICOM images.

For the single-view Varian TrueBeam case, we analytically more » derived them from the raw patient-independent PS data in IAEA’s database, partial geometry information of the jaw and MLC as well as the fluence map. For the helical Tomotherapy case, the PS data were calculated in our previous study (Su et al. Methods: The patient-specific source of Tomotherapy and Varian TrueBeam Linacs was modeled using the PS approach. Purpose: (1) To perform phase space (PS) based source modeling for Tomotherapy and Varian TrueBeam 6 MV Linacs, (2) to examine the accuracy and performance of the ARCHER Monte Carlo code on a heterogeneous computing platform with Many Integrated Core coprocessors (MIC, aka Xeon Phi) and GPUs, and (3) to explore the software micro-optimization methods. Together with hand-tuned assembly kernels and state-of-the-art parallelization, this provides extremely high performance and cost efficiency for high-throughput as well as massively parallel simulations.

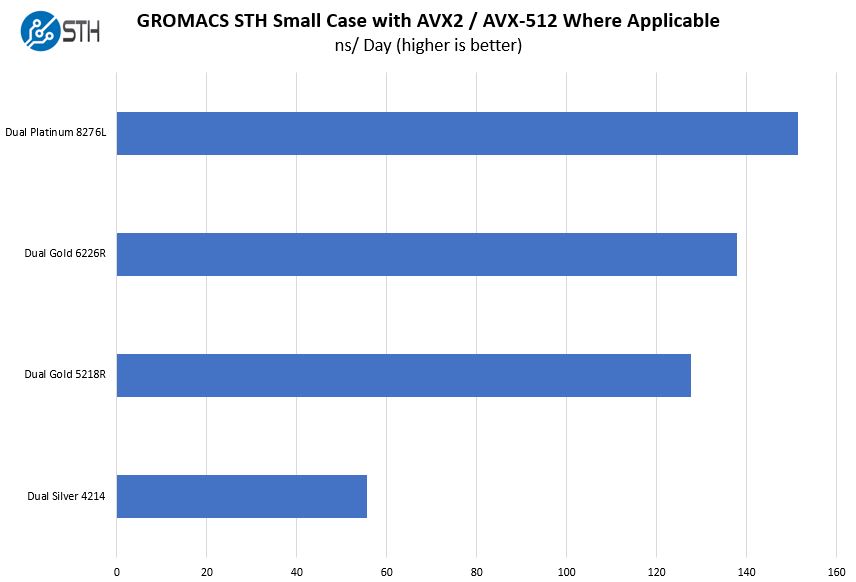
GROMACS supports several implicit solvent models, as well as new free-energy algorithms, and the software now uses multithreading for efficient parallelization even on low-end systems, including windows-based workstations. The software now automatically handles wide classes of biomolecules, such as proteins, nucleic acids and lipids, and comes with all commonly used force fields for these molecules built-in. As more » a result, we present a range of new simulation algorithms and features developed during the past 4 years, leading up to the GROMACS 4.5 software package. These applications share a need for fast and efficient software that can be deployed on massive scale in clusters, web servers, distributed computing or cloud resources. At the same time, advances in performance and scaling now make it possible to model complex biomolecular interaction and function in a manner directly testable by experiment. In this study, molecular simulation has historically been a low-throughput technique, but faster computers and increasing amounts of genomic and structural data are changing this by enabling large-scale automated simulation of, for instance, many conformers or mutants of biomolecules with or without a range of ligands.


 0 kommentar(er)
0 kommentar(er)
